| Table 1. Primer sequence information. | |||||||
| Locus name | Primer sequence (5’ to 3’) | Reference | Label | GenBank accession number | Expected size range | Amplified size range | Concentration of primers in multiplex PCR (µM) |
| WGA 001 | F: ATTGGAAGGGAAGGGAAATG R: CGCGCACATACGTAAATCAC | Dangel et al. 2005 | 6 FAM | AY465952 | 180–192 | 186–192 | 0.3 |
| WGA 004 | F: TGTTGCATTGACCCACTTGT R: TAAGCCAACATGGTATGCCA | Dangel et al. 2005 | NED | AY465953 | 228–238 | 228–230 | 0.15 |
| WGA 005 | F: CAGTTTGTCCCACACCTCCT R: AACCCATGGTGAGAGTGAGC | Woeste et al. 2002 | 6 FAM | No data | 240–266 | Multiallele pattern | - |
| WGA 009 | F: CATCAAAGCAAGCAATGGG R: CCATTGCTCTGTGATTGGG | Dangel et al. 2005 | VIC | AY465954 | 229–252 | 236–244 | 0.15 |
| WGA 027 | F: AACCCTACAACGCCTTGATG R: TGCTCAGGCTCCACTTCC | Woeste et al. 2002 | 6 FAM | AY333951 | 192–210 | 202–208 | - |
| WGA 032 | F: CTCGGTAAGCCACACCAATT R: GGACCCAGCTCCTCTTCTCT | Woeste et al. 2002 | VIC | AY333952 | 120–196 | Multiallele pattern | - |
| WGA 071 | F: ACCCGAGAGATTTCTGGGAT R: GGACCCAGCTCCTCTTCTCT | Woeste et al. 2002 | 6 FAM | No data | 136–210 | 205–207 | - |
| WGA 069 | F: TTAGATTGCAAACCCACCCG R: AGATGCACAGACCAACCCTC | Dangel et al. 2005 | PET | AY333953 | 158–180 | 164–184 | 0.15 |
| WGA 089 | F: ACCCATCTTTCACGTGTGTG R: TGCCTAATTAGCAATTTCCA | Dangel et al. 2005 | 6 FAM | AY352440 | 212–220 | 212–220 | 0.25 |
| WGA 118 | F: TGTGCTCTGATCTGCCTCC R: GGGTGGGTGAAAAGTAGCAA | Dangel et al. 2005 | NED | AY479958 | 184–209 | 184–206 | 0.1 |
| WGA 148 | F: GGTGAACTCCCATAGGGGTA R: CCAATGCTACTTGCAGAACC | Woeste et al. 2006 | VIC | DQ307431 | 253–271 | 252–296 | 0.2 |
| WGA 202 | F: CCCCATCTACCGTTGCACTTT R: GCTGGTGGTTCTATCATGGG | Dangel et al. 2005 | VIC | AY479959 | 238–275 | 251–275 | - |
| WGA 204 | F: GGGTCTCGCCTTCTTTTCTT R: CACAGAGAGAAGCACGGGTA | Woeste et al. 2007 | NED | DQ307432 | 172–178 | 174–176 | 0.1 |
| WGA 225 | F: AATCCCTCTCCTGGGCAG R: TGTTCCACTGACCACTTCCA | Dangel et al. 2005 | PET | AY479960 | 190–202 | 192–204 | - |
| WGA 256 | F: TGAAGACAACAAAACTGCGC R: CCGGCATTGTTTCTGAAAAT | Woeste et al. 2009 | PET | DQ307432 | 227–253 | 249–291 | 0.2 |
| WGA 276 | F: CTCACTTTCTCGGCTCTTCC R: GGTCTTATGTGGGCAGTCGT | Dangel et al. 2005 | 6 FAM | AY479961 | 143–192 | 168–194 | - |
| WGA 321 | F: TCCAATCGAAACTCCAAAGG R: GTCCAAAGACGATGATGGA | Dangel et al. 2005 | NED | AY479962 | 222–225 | 223–239 | - |
| WGA 331 | F: TCCCCCTGAAATCTTCTCCT R: CGGTGGTGTAAGGCAAATG | Dangel et al. 2005 | VIC | AY479963 | 273–277 | 271–275 | - |
| WGA 332 | F: ACGTCGTTCTGCACTCCTCT R: GCCACAGGAACGAGTGCT | Foroni et al. 2006 | PET | AY479964 | 214–225 | 215–225 | 0.1 |
| WGA 349 | F: GTGGCGAAAGTTTATTTTTTGC R: ACAAATGCACAGCAGCAAAC | Woeste et al. 2002 | 6 FAM | AY479965 | 258–274 | 262–274 | 0.25 |
| WGA 376 | F: GCCCTCAAAGTGATGAACGT R: TCATCCATATTTACCCCTTTCG | Woeste et al. 2002 | NED | AY479966 | 218–254 | 230–252 | - |
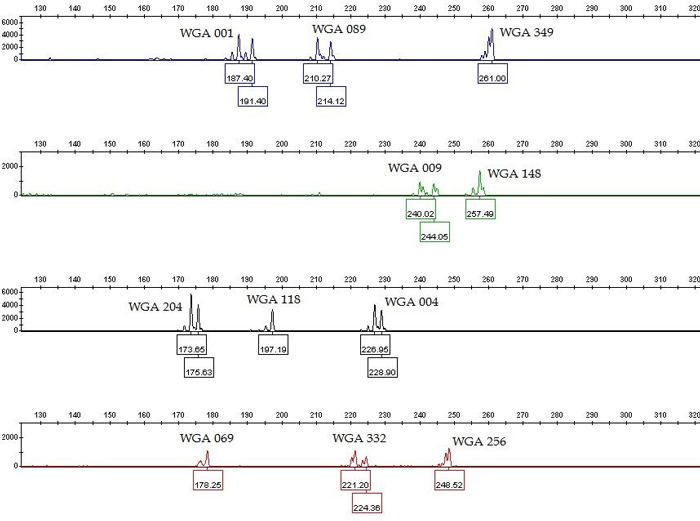
Fig. 1. Genotype profile (electropherogram) of the eleven microsatellite loci (SSR markers) of common walnut generated by multiplex PCR. The x-axis represents DNA fragment size in base pairs, and the y-axis represents fluorescence units. The SSR profile is displayed in the blue (6-FAM labeled primers), green (VIC labeled primers), yellow (NED labeled primers) and red (PET labeled primers) channels of a five-color fluorescent system, with the orange (LIZ labeled fragments) channel being used for a size marker (not shown). View larger in new window/tab.
| Table 2. Gene diversity parameters of eleven common walnut SSR markers, sample size (N), number of observed alleles (Na), effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), fixation index (FIS), and Hardy-Weinberg equilibrium (HW). | |||||||
| Locus | N | Na | Ne | Ho | He | FIS | HW |
| WGA 001 | 15 | 4 | 2,514 | 0,200 | 0,602 | 0,668 | *** |
| WGA 004 | 15 | 2 | 1,923 | 0,533 | 0,480 | –0,111 | ns |
| WGA 009 | 15 | 3 | 2,761 | 0,533 | 0,638 | 0,164 | ns |
| WGA 069 | 15 | 7 | 4,091 | 0,333 | 0,756 | 0,559 | * |
| WGA 089 | 15 | 4 | 3,103 | 0,667 | 0,678 | 0,016 | ns |
| WGA 118 | 15 | 4 | 2,571 | 0,667 | 0,611 | –0,091 | ns |
| WGA 148 | 15 | 6 | 2,586 | 0,400 | 0,613 | 0,348 | *** |
| WGA 204 | 15 | 2 | 1,069 | 0,067 | 0,064 | –0,034 | ns |
| WGA 256 | 15 | 4 | 2,432 | 0,667 | 0,589 | –0,132 | ns |
| WGA 332 | 15 | 3 | 2,761 | 0,667 | 0,638 | –0,045 | ns |
| WGA 349 | 15 | 3 | 2,381 | 0,467 | 0,580 | 0,195 | ns |
| Mean | 15 | 3,818 | 2,563 | 0,473 | 0,568 | 0,140 | |
| SE | 0 | 0,464 | 0,222 | 0,062 | 0,054 | 0,084 | |
| ns = not significant, * P<0.05, ** P<0.01, *** P<0.001 | |||||||