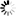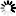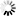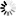Genetic patterns in Pinus nigra from the central Balkans inferred from plastid and mitochondrial data
Šarac Z., Dodoš T., Rajčević N., Bojović S., Marin P., Aleksić J. (2015). Genetic patterns in Pinus nigra from the central Balkans inferred from plastid and mitochondrial data. Silva Fennica vol. 49 no. 5 article id 1415. https://doi.org/10.14214/sf.1415
Highlights
- Seven populations of Pinus nigra from the central Balkans, representing four infraspecific taxa, were analyzed with chloroplast microsatellites and a mitochondrial locus
- Molecular data failed to support infraspecific circumscriptions
- Levels of genetic diversities/differentiation at both genomes were in the range of those reported in western Mediterranean populations of P. nigra
- Iberian/African and Balkans’ populations share one mtDNA polymorphism and differ in three mutations.
Abstract
Pinus nigra J.F. Arnold, European black pine, is a typical component of Mediterranean and sub-Mediterranean coniferous forests with highly fragmentary distribution. Western Mediterranean populations of this species have been studied genetically to date, while eastern populations from the central Balkans, which are larger and more abundant, are still genetically understudied. We analyzed seven populations of P. nigra representing all infraspecific taxa recognized within the central Balkans (subspecies nigra with varieties nigra and gocensis Đorđević; and subspecies pallasiana (Lamb.) Holmboe with varieties pallasiana and banatica (Endl.) Georgescu et Ionescu), with three chloroplast microsatellites (cpDNA SSRs) and one mitochondrial (mtDNA) locus. Although our molecular data failed to support circumscription of studied infraspecific taxa, we found that genetic patterns at both genomes are in accordance with those found previously in westward populations of this species, that is – exceptionally high levels of genetic diversity (HT = 0.949) and low genetic differentiation (GST = 0.024) at the cpDNA level, and moderate levels of genetic diversity (HT = 0.357) and genetic differentiation (GST = 0.358) at the mtDNA level. Based on genealogical relations of mtDNA types currently present in Balkans’ and Iberian/African populations, we inferred that the ancestral gene pool of P. nigra already harbored polymorphism at position 328 prior to the divergence to two lineages currently present in westward and eastward parts of the species range distribution. Subsequent occurrence of three mutations, which distinguish these two lineages, suggests their long-term isolation.
Keywords
genetic differentiation;
genetic diversity;
the Balkans;
European black pine;
chloroplast microsatellites;
mitochondrial nad7 intron 1;
haplotype network
-
Šarac,
University of Niš, Faculty of Sciences and Mathematics, Department of Biology and Ecology, Višegradska 33, 18000 Niš, Serbia
E-mail
saraczorica@gmail.com

- Dodoš, University of Belgrade, Faculty of Biology, Institute of Botany and Botanical Garden “Jevremovac”, Studentski trg 16, 11000 Belgrade, Serbia E-mail tanjadodos@bio.bg.ac.rs
- Rajčević, University of Belgrade, Faculty of Biology, Institute of Botany and Botanical Garden “Jevremovac”, Studentski trg 16, 11000 Belgrade, Serbia E-mail nemanja@bio.bg.ac.rs
- Bojović, University of Belgrade, Institute for Biological Research “Siniša Stanković”, Boulevard Despota Stefana 142, 11060 Belgrade, Serbia E-mail bojovic@ibiss.bg.ac.rs
- Marin, University of Belgrade, Faculty of Biology, Institute of Botany and Botanical Garden “Jevremovac”, Studentski trg 16, 11000 Belgrade, Serbia E-mail pdmarin@bio.bg.ac.rs
- Aleksić, University of Belgrade, Institute of Molecular Genetics and Genetic Engineering, Vojvode Stepe 444a, P.O. Box 23, 11000 Belgrade, Serbia E-mail aleksic_jelena@yahoo.com.au
Received 30 June 2015 Accepted 4 September 2015 Published 18 September 2015
Views 126492
Available at https://doi.org/10.14214/sf.1415 | Download PDF

1 Introduction
Pinus nigra J.F. Arnold, European black pine, is one of the economically and ecologically most important pine species in Europe. It is a typical component of Mediterranean and sub-Mediterranean coniferous forests, with highly fragmentary distribution extending from the North Africa, the northern Mediterranean Basin and eastwards to the Black Sea (Gaussen et al. 1993). Due to its Tertiary origin, relict nature and a prominent phenotypic plasticity throughout the range distribution, its taxonomic status and infraspecific classification are still somewhat unclear (e.g. Vidaković 1991; Gaussen et al. 1993; Christensen 1993; see also references in Naydenov et al. 2006 and Ganopoulos et al. 2013).
To date, fragmentary distributed natural populations of P. nigra from the western part of the species range have been studied genetically (e.g. Afzal-Rafii and Dodd 2007; Jaramillo-Correa et al. 2010; Soto et al. 2010). However, those from the Balkans and Turkey, which are larger and more abundant, have occasionally been studied from various aspects (e.g. Liber et al. 2003; Lučić et al. 2010; 2013; Gülsoy et al. 2014) but rarely from a population genetics aspect (e.g. Naydenov et al. 2006, 2015; Šarac et al. 2015). The genetic knowledge on these populations is required not only for shedding new light on species infraspecific circumscription and overall genetic and/or phylogeographic structure, but also for implementation of suitable conservation measures and future breeding efforts (Frankham et al. 2002). Studies of this kind, however, are not straight-forward because, on the one hand, P. nigra is one of the most commonly planted conifers in Europe and the origin of the planted material cannot be tracked in all cases, and, on the other hand, this predominantly outcrossing species is characterized by a rather high dispersal ability (pollination and seed dispersal by wind, Afzal-Rafii and Dodd 2007; Jaramillo-Correa et al. 2010; Soto et al. 2010) suggesting possible introgression of alien gene pools into native populations.
Within the central Balkans, two subspecies of P. nigra (ssp. nigra and ssp. pallasiana (Lamb.) Holmboe), and their varieties (var. nigra and gocensis Đorđević of the former subspecies, and var. pallasiana and banatica (Endl.) Georgescu et Ionescu of the latter subspecies), have been recognized (Jovanović 1992). It has been shown recently that these infraspecific taxa differ at the biochemical level (Bojović et al. 2012; Šarac et al. 2013). In the frame of the present study, we employ: 1) paternally inherited and potentially more variable chloroplast microsatellites (cpDNA SSRs) shown to be informative for assessing genetic patterns in P. nigra and other pines (Naydenov et al. 2006; Afzal-Rafii and Dodd 2007; Jaramillo-Correa et al. 2010; Soto et al. 2010), and 2) a more slowly evolving maternally inherited mitochondrial (mtDNA) locus nad7 intron 1, proven to be suitable for depicting phylogeographic structure in westward populations of P. nigra (Jaramillo-Correa et al. 2010), and test them in individuals of P. nigra from seven populations representing all recognized infraspecific taxa of this species from the central Balkans in order to: 1) assess whether cpDNA and/or mtDNA markers are suitable for distinguishing infraspecific taxa of P. nigra from the central Balkans; 2) estimate levels of genetic diversity and genetic structuring at cpDNA and mtDNA levels in these genetically understudied populations of P. nigra; and 3) use available variation at the studied mtDNA locus to make inferences on historical events that have led to the current genetic patterns in westward and eastward parts of the species range.
2 Material and methods
2.1 Plant material and DNA extraction
Samples (young twigs with needles) were collected in early fall 2009 from 14 – 15 trees per population from seven native populations of P. nigra from the central Balkans which represent all infraspecific taxa of this species (ssp. nigra represented by var. nigra and var. gocensis; and ssp. pallasiana represented by var. pallasiana and var. banatica) recognized by the Flora of Serbia (Jovanović 1992). We sampled 104 trees in total (Table 1). Trees sampled from each population were distant at least 30 m from each other, and sampled populations were not in the vicinity of known planted trees/populations of this species. Plant material, deposited in labeled polyethylene bags, was stored at –20 °C prior to DNA extraction.
| Table 1. The infraspecific taxa of Pinus nigra from the central Balkans used in the present study, characteristics of sampling localities, and estimates of chloroplast microsatellite (cpDNA SSR) and mitochondrial (mtDNA) diversity. View in new window/tab. |
Approximately 30 mg of the plant tissue of each individual, homogenized with TissueLyser II (Qiagen, Valencia, CA, USA), was used for DNA extraction following Aleksić et al. (2012).
2.2 Molecular markers
Nine chloroplast (cpDNA) microsatellites (Pt1254, Pt26081, Pt36480, Pt45002, Pt71936, Pt79951, Pt15169, Pt30204, and Pt87268, Vendramin et al. 1996) were initially tested in a panel of seven randomly selected individuals from each out of seven populations following PCR amplification procedure of Vendramin et al. (1996) and using a peqStar 96 Universal thermal cycler (PEQLAB Biotechnologie GmbH, Erlangen, Germany). Three loci, Pt15169, Pt30204, and Pt87268, showing a successful amplification in all individuals and a potential length polymorphism of PCR products upon separation by 2% agarose gel electrophoresis, were further used and PCR amplified using fluorescently labelled forward (F) primers (DS-33, Applied Biosystems, USA). PCR products were separated comercially via capillary electrophoresis by Macrogen Europe, Amsterdam, the Netherlands (http://dna.macrogen.com/eng/) using 96-capillary 3730xl DNA Analyzer automated sequencer (Applied Biosystems, Inc. USA). Sizing of fragments was performed manually with GeneMapper ver. 4.0 (Applied Biosystems, Foster City, USA).
Three mitochondrial (mtDNA) loci, the second intron of the NADH dehydrogenase subunit 1 gene - nad1 gene intron 2 (primers published by Demesure et al. 1995), nad5 intron 4 (Dumolin-Lapegue et al. 1997), and nad7 intron 1 (Jaramillo-Correa et al. 2004; Tian et al. 2010), were initially tested in the same panel of seven P. nigra individuals following PCR conditions provided in the corresponding publications. However, PCR products were obtained only at locus nad7 intron 1 using the F primer of Tian et al. (2010) and the R primer of Jaramillo-Correa et al. (2004), and the following PCR conditions: a total volume of 25 μl containing 25 ng template DNA, 1 × Taq buffer with (NH4)2SO4 (Fermentas UAB, Vilnius, Lithuania), 2.5 mM MgCl2 (Fermentas UAB, Vilnius, Lithuania), 0.2 mM dNTPs, 0.1 μM of each forward and reverse primers, 0.80% BSA (Bovine Serum Albumin, Fermentas UAB, Vilnius, Lithuania), and 0.025 U Taq DNA polymerase (Fermentas UAB, Vilnius, Lithuania), and a touchdown amplification protocol: 4 min initial denaturation at 94 °C, 11 cycles of 1 min denaturation at 94 °C, 45 s annealing at 68 °C, 2 min extension at 72 °C with progressive decrease of the annealing temperature by 0.5 °C/cycle, followed by 26 cycles of 1 min denaturation at 94 °C, 45 s annealing at 56 °C, 2 min extension at 72 °C, and final extension of 10 min at 72 °C. PCR products were Sanger sequenced by Macrogen Europe using F or both (F and R) primers (see Results).
2.3 Data analysis
At the cpDNA level, we assessed the following parameters of genetic diversity at the population level and in overall sample: the number of alleles (A), the number of private alleles (PA), the number of chloroplast haplotypes (ch), the number of private haplotypes (cph), unbiased within population gene diversity (HS, equivalent to HE for diploid data, Weir, 1996), and the total gene diversity (HT) using CONTRIB 1.02 (Petit et al. 1998). Using the same software, we assessed also the number of mitochondrial haplotypes (mh), HS and HT at the mtDNA level in populations and at the species level. The standardized coefficients of genetic differentiation among populations, GST, at the cpDNA and mtDNA levels were also calculated in CONTRIB.
Pairwise population differentiation estimates, FST, based on cpDNA haplotype frequencies, and the analysis of molecular variance (AMOVA, Excoffier et al. 1992), based on the sum of squared number of repeat difference between cpDNA haplotypes (Slatkin 1995), were calculated using ARLEQUIN 3.11 (Excoffier et al. 2005). The significance of tests was examined by 15 000 permutations.
TESS 1.2 (François et al. 2006; Chen et al. 2007) was used to further study genetic differentiation of populations based on cpDNA data, by estimating the number of genetically similar clusters. TESS incorporates the spatial locations of all individuals into the analyses to assess genetic cluster membership. Spatial coordinates of individual trees within defined areas, corresponding to population sizes, were generated using population sample coordinates. Using no admixture model and interaction parameter of 0.6, 100 runs at each K ranging from 2 to 4 with 50 000 sweeps and a burn-in of 10 000 for each run were performed. At each K, 20 runs with the lowest deviance information criterion (DIC) were recorded and used for calculating average DICs. As recommended by Durand et al. (2009), the optimal number of clusters may be determined by plotting average DIC against K, and depicting K at the beginning of the plateau.
MUSCLE (Edgar 2004) implemented in MEGA 5.04 (Tamura et al. 2011) was used for the alignment of mtDNA sequences, which were checked and edited manually. Genealogical relationships among mtDNA haplotypes were assessed by constructing median-joining network with NETWORK 4.6.1.2 (Bandelt et al. 1999). For this analysis, we used also nad7 intron 1 sequences of P. nigra available in the GenBank (accessed on 01-12-2014): accessions HQ185286, HQ185287, HQ185288, and HQ185289 (Jaramillo-Correa et al. 2010), and sequences of P. sylvestris L. used as outgroup (DQ665913, DQ665914, and DQ665915, Naydenov et al. 2007). Sequence gaps were treated as a fifth character state after recoding insertions/deletions (indels) longer than 1 base-pair (bp) as single-bp indels.
3 Results
Three cpDNA SSRs were successfully amplified in 104 P. nigra individuals, yielding 5–9 alleles per locus, 22 alleles in total, and 38 haplotypes (Table 1). The number of PA and cph were high (7 and 22, respectively) as well as average HS (0.926) and HT (0.949). Genetic differentiation of populations was low (GST = 0.024). MtDNA locus nad7 intron 1 was successfully amplified and sequenced with F primer in 99 P. nigra individuals while sequencing with R primer was successful in 73 individuals. The distribution of mtDNA haplotypes among populations was not random, and two detected haplotypes (T and G, see later) were found at more or less equal frequencies in populations of P. nigra ssp. nigra var. nigra (I, II and III), while populations IV to VII, representing the other studied taxa, were fixed for haplotype T. Thus, the average HS and HT were low to moderate (0.229 and 0.357, respectively), while GST was moderate (0.358).
All population pairwise FST values were insignificant, and ranged from 0.066 (populations IV and V representing P. nigra ssp. nigra var. gocensis) to 0.000 or were negative (data not shown). Although AMOVA revealed that 1.72% and 98.28% of overall molecular variation were attributed to among and within population variation, respectively, these values were insignificant. When two subspecies of P. nigra were compared (populations I to V representing ssp. nigra vs. populations VI and VII of ssp. pallasiana), somewhat increased among population variation (2.06%), decreased within population variation (97.20%) and 0.73% of variation among populations within groups were observed. All values, however, were insignificant. The lack of genetic structuring of populations at the cpDNA level was revealed by TESS analysis as well, because at each tested K, all individuals were strongly assigned to a single gene pool, with a few individuals from populations VI and VII assigned to alternative gene pools (Fig. 1d). Therefore, calculations required for assessing the optimal number of clusters were not performed because individuals from all populations belong to a single gene pool.
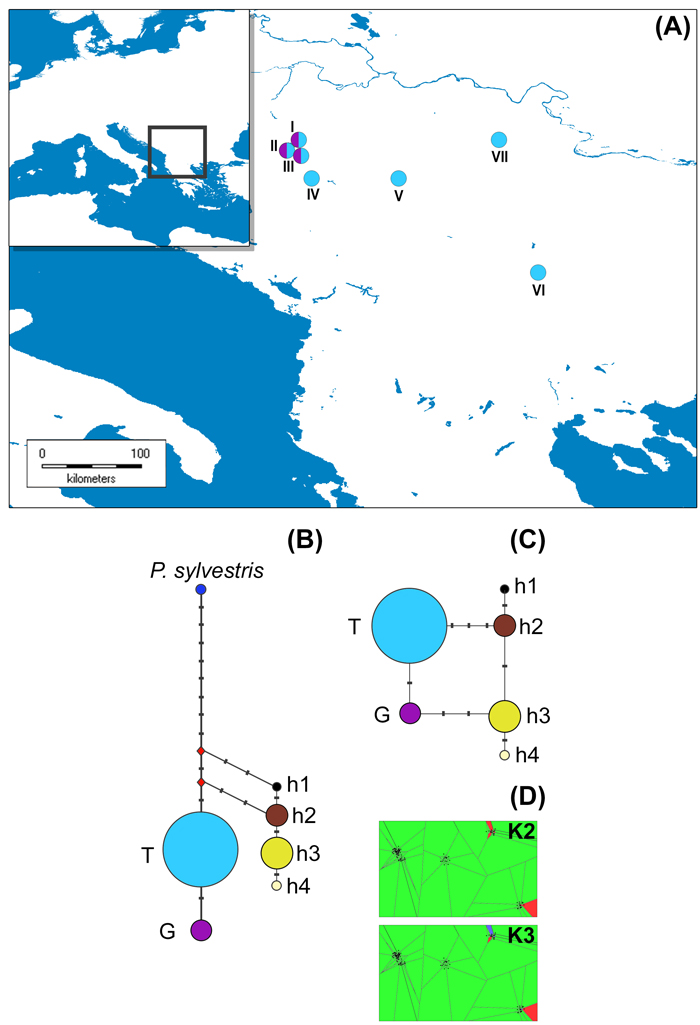
Fig. 1. The distribution of studied P. nigra populations within the central Balkans and the abundance of mtDNA haplotypes (A), haplotype networks based on mitochondrial data (B and C) and outcomes of TESS analyses based on chloroplast data (D).
Populations I, II and III represent P. nigra ssp. nigra var. nigra, populations IV and V represent P. nigra ssp. nigra var. gocensis, population VI represents P. nigra ssp. pallasiana var. pallasiana, and population VII represents P. nigra ssp. pallasiana var. banatica (see also Table 1); the size of circles in haplotype network rooted with P. sylvestris (B) and unrooted haplotype network (C) is proportional to the relative frequency of haplotypes T and G in eastward populations (see Table 1), and haplotypes h1 to h4 in westward populations, but the relative frequency of haplotypes in these regions is not comparable; mtDNA haplotype labels are given in the legend of Table 2; D – clustering of populations at K = 2 and K = 3 using TESS.
The length of aligned matrices comprising nad7 intron 1 sequences of P. nigra generated in the present study, and those reported by Jaramillo-Correa et al. (2010), as well as sequences of P. sylvestris (Naydenov et al. 2007), were 985 bp, 985 bp and 991 bp, respectively. The positions of variable sites (point and length mutations) in the latter matrix are shown in Table 2. P. nigra mtDNA haplotypes from the Balkans harbored a single polymorphic site at position 328 (G/T transversion), yielding two mtDNA haplotypes: T (harboring T at position 328), and G (G at position 328), which were deposited in the GenBank (accessions KT343352 and KT343353). The Balkans’ and Iberian/African mtDNA haplotypes of P. nigra shared variability at position 328, and differed in three mutations, at positions 435, 472 and 520 (Table 2). Nucleotide characters G and A at positions 435 and 520, respectively, are synapomorphic for individuals from Iberia/Africa, while a single-bp insertion (C) at position 472 is synapomorphic for individuals from the central Balkans. All P. sylvestris sequences harbored T at position 328, and differed from P. nigra sequences in 8 mutations, at positions 334, 337, 396, 445, 565, 775, 889, and 931 (see Table 2).
| Table 2. Positions of variable sites in 991 bp long aligned mtDNA nad7 intron1 matrix comprised of six Pinus nigra haplotypes and three Pinus sylvestris haplotypes used as an outgroup. View in new window/tab. |
Haplotype networks constructed with and without outgroup, P. sylvestris, are shown in Fig. 1b and 1c, respectively.
4 Discussion
High levels of cpDNA SSR diversity (HT = 0.949) and low genetic differentiation of populations (GST = 0.024) found in populations of P. nigra from the central Balkans are in the range of those detected in westward populations of this species (Afzal-Rafii and Dodd 2007; Jaramillo-Correa et al. 2010; Soto et al. 2010). These findings, along with upper Miocene fossil records of P. nigra in central and southern Serbia (Petković 1987), would suggest a long persistence of this species within the Balkans, supported also by findings of Naydenov et al. (2006). Interestingly, these authors found somewhat lower levels of cpDNA diversity and increased genetic differentiation of P. nigra populations in Bulgaria, with four groups of populations whose distinctiveness was explained by historical events, specific topography of the region and anthropogenic impact. The alternative clustering of these populations found at the nuclear genome, and discordance of cpDNA and nuclear data, was attributed to different migration patterns and effective radius of dispersal of P. nigra pollen and seed (Naydenov et al. 2015). On the other hand, we found moderately high levels of mtDNA diversity in eastward populations of P. nigra (HT = 0.357), and a non-random distribution of two mtDNA haplotypes, which has led to the moderate levels of genetic differentiation (GST = 0.358). Since the observed levels of genetic differentiation at the mtDNA level are usually expected at much larger studied areas than ours (e.g. Jaramillo-Correa et al. 2010; Tian et al. 2010; Aleksić and Geburek 2014 and references therein), we assumed that the Tara Mountain, with P. nigra populations characterized by increased levels of mtDNA diversity, may represent an interglacial refugium of this species, similarly as observed in another cold-adapted species, Picea omorika (Panč.) Purk. (Aleksić and Geburek 2014).
Although it has been shown recently that cpDNA variation in closely related spruces is more species-specific than mtDNA variation, and that the latter is often shared among related conifer species (Du et al. 2009), we were not able to use cpDNA nor mtDNA polymorphisms to support infraspecific circumscription of studied P. nigra taxa which has recently been implied at the biochemical level (Bojović et al. 2012; Šarac et al. 2013). The sole conclusions with regard to this aspect of our study are that at the cpDNA level, two studied populations of P. nigra ssp. nigra var. gocensis display the highest but insignificant genetic differentiation, and that populations VI and VII, representing varieties pallasiana and banatica of P. nigra ssp. pallasiana, respectively, may harbor genetically distinct individuals. It is worth mentioning that these populations are at the range-edge of P. nigra ssp. pallasiana distribution (Vidaković 1991) and thus, may be genetically different from populations of these taxa found within their main range (e.g. Bjedov et al. 2015 and references therein). At the mtDNA level, populations of P. nigra ssp. nigra var. nigra are genetically diverse while populations of the other studied infraspecific taxa are invariable. Nonetheless, mtDNA variation enabled new insights into historical events that have led to the current distribution of mtDNA diversity in westward and eastward parts of the P. nigra range.
Based on the haplotype network rooted with P. sylvestris (Fig. 1b), we may assume that P. nigra mtDNA haplotypes harboring G at position 328 (Table 2), which are currently less abundant in eastward but prevalent in westward populations (h3 and h4), are derived. This would imply that the ancestral gene pool of P. nigra, which initially spread throughout the Mediterranean Basin, harbored T at position 328, and that the same mutation (T → G at position 328) must have had occur twice, i.e. in the ancestral P. nigra gene pool within the Balkans, and in the ancestral gene pool of this species within the western Mediterranean Basin. Alternatively, due to the long persistence of P. nigra in the Mediterranean (Naydenov et al. 2006, 2015; Jaramillo-Correa et al. 2012) and its high dispersal ability, it is possible that the above mentioned mutation occurred at one site, and expanded later on to the other site. However, the more plausible and parsimonious scenario is that the polymorphism at position 328 was already present in the ancestral gene pool of P. nigra which inhabited the Mediterranean Basin, and that three mutations, which distinguish two lineages of this species confined to the Balkans and the Iberian Peninsula/northern Africa, occurred after their divergence. It is worth mentioning the nucleotide characters G and A at positions 435 and 520, respectively, and one bp insertion (C) at position 472 may be used as molecular diagnostic characters for distinguishing westward and eastward P. nigra lineages. These findings altogether further support long persistence of P. nigra in southern Europe, and long-term isolation of two lineages of this species detected in the present study.
Acknowledgement
This research was supported by grants 173029 and 173030 from the Ministry of Education, Science, and Technology of the Republic of Serbia.
References
Afzal-Rafii Z., Dodd R.S. (2007). Chloroplast DNA supports a hypothesis of glacial refugia over postglacial recolonization in disjunct populations of black pine (Pinus nigra) in western Europe. Molecular Ecology 16: 723–736. http://dx.doi.org/10.1111/j.1365-294X.2006.03183.x.
Aleksić J.M., Geburek T. (2014). Quaternary population dynamics of an endemic conifer, Picea omorika, and their conservation implications. Conservation Genetics 15: 87–107. http://dx.doi.org/10.1007/s10592-013-0523-6.
Aleksić J.M., Stojanović D., Banović B., Jančić R. (2012). A simple and efficient DNA isolation method for Salvia officinalis. Biochemical Genetics 50: 881–892. http://dx.doi.org/10.1007/s10528-012-9528-y.
Bandelt H.-J., Forster P., Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48.
Bjedov I., Obratov-Petković D., Mišić D., Šiler B., Aleksić J.M. (2015). Genetic patterns in range-edge populations of Vaccinium species from the central Balkans: implications on conservation prospects and sustainable usage. Silva Fennica 49(4) article 1283. http://dx.doi.org/10.14214/sf.1283.
Bojović S., Šarac Z., Nikolić B., Tešević V., Todosijević M., Veljić M., Marin P.D. (2012). Composition of n-alkanes in natural populations of Pinus nigra from Serbia – chemotaxonomic implications. Chemistry and Biodiversity 9: 2761–2774. http://dx.doi.org/10.1002/cbdv.201200051.
Chen C., Durand E., Forbes F., Francois O. (2007). Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Molecular Ecology Notes 7: 747–756. http://dx.doi.org/10.1111/j.1471-8286.2007.01769.x.
Christensen K.I. (1993). Comments on the earliest validly published varietal name for the Corsican pine. Taxon 42: 649–653. http://dx.doi.org/10.2307/1222544.
Demesure B., Sodzi N., Petit R.J. (1995). A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology 4: 129–131. http://dx.doi.org/10.1111/j.1365-294X.1995.tb00201.x.
Dumolin-Lapegue S., Pemonge M.-H., Petit R.J. (1997). An enlarged set of consensus primers for the study of organelle DNA in plants. Molecular Ecology 6: 393–397. http://dx.doi.org/10.1046/j.1365-294X.1997.00193.x.
Durand E., Chen C., François O (2009). TESS version 2.3 reference manual. http://membres-timc.imag.fr/Olivier.Francois/manual.pdf. [Cited Jun 2012].
Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. http://dx.doi.org/10.1093/nar/gkh340.
Excoffier L., Smouse P.E., Quattro J.M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491.
Excoffier L., Laval G., Schneider S. (2005). Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50.
François O., Ancelet S., Guillot G. (2006). Bayesian clustering using hidden Markov random fields in spatial population genetics. Genetics 174: 805–816. http://dx.doi.org/10.1534/genetics.106.059923.
Frankham R., Ballou. J, Briscoe D. (2002). Introduction to Conservation Genetics. Cambridge University Press, New York.
Ganopoulos I., Aravanopoulos F., Madesis P., Pasentsis K., Bosmali I., Ouzounis C., Tsaftaris A. (2013). Taxonomic identification of Mediterranean pines and their hybrids based on the high resolution melting (HRM) and trnL approaches: from cytoplasmic inheritance to timber tracing. PLoS ONE 8: e60945. http://dx.doi.org/10.1371/journal.pone.0060945.
Gaussen H., Heywood V.H., Charter A.O. (1993). Pinus L. In: Tutin T.G., Heywood V.H., Burges N.A., Valentine D.H., Walters S.M., Webb B.A. (eds.). Flora Europaea, Vol. 1. Cambridge University Press, Cambridge. p. 40–44.
Gülsoy A.D., Gülsoy A.M., Çengel B., Kaya Z. (2014). The evolutionary divergence of Pinus nigra subsp. pallasiana and its varieties based on noncoding trn regions of chloroplast genomes. Turkish Journal of Botany 38: 627–636. http://dx.doi.org/10.3906/bot-1312-26.
Jaramillo-Correa J.P., Beaulieu J., Bousquet J. (2004). Variation in mitochondrial DNA reveals multiple distant glacial refugia in black spruce (Picea mariana), a transcontinental North American conifer. Molecular Ecology 13: 2735–2747. http://dx.doi.org/10.1111/j.1365-294X.2004.02258.x.
Jaramillo-Correa J.P., Grivet D., Terrab A., Kurt Y., De-Lucas A.I., Wahid N., Vendramin G.G., González-Martínez S.C. (2010). The Strait of Gibraltar as a major biogeographic barrier in Mediterranean conifers: a comparative phylogeographic survey. Molecular Ecology 19: 5452–5468. http://dx.doi.org/10.1111/j.1365-294X.2010.04912.x.
Jovanović B. (1992). P. nigra Arn. In: Sarić M. (ed.). The Flora of Serbia, Vol. 1. SANU, Belgrade. p. 200–202.
Liber Z., Nikolić T., Mitić B., Šatović Z. (2003). RAPD markers and black pine (Pinus nigra Arnold) intraspecies taxonomy - evidence from the study of nine populations. Acta Societatis Botanicorum Poloniae 72: 249–257. http://dx.doi.org/10.5586/asbp.2003.033.
Lučić A., Mladenović-Drinić S., Stavretović N., Isajev V., Lavadinović V., Rakonjac L., Novaković M. (2010). Genetic diversity of Austrian pine (Pinus nigra Arnold) populations in Serbia revealed by RAPD. Archives of Biological Science 62: 329–336.
Lučić A., Cvjetićanin R., Novaković-Vuković M., Ristić D., Popović V., Rakonjac L., Mladenović Drinić S. (2013). Interpopulation variability of Austrian pine (Pinus nigra Arnold) in Serbia. Genetika 45: 641–654. http://dx.doi.org/10.2298/GENSR1303641L.
Naydenov K., Senneville S., Beaulieu J., Tremblay F., Bousquet J. (2007). Glacial vicariance in Eurasia: mitochondrial DNA evidence from Scots pine for a complex heritage involving genetically distinct refugia at mid-northern latitudes and in Asia Minor. BMC Evolutionary Biology 7: 233–244. http://dx.doi.org/10.1186/1471-2148-7-233.
Naydenov K.D., Tremblay F., Fenton N., Alexandrov A. (2006). Structure of Pinus nigra Arn. populations in Bulgaria revealed by chloroplast microsatellites and terpenes analysis: provenance tests. Biochemical Systematics and Ecology 34: 562–574. http://dx.doi.org/10.1016/j.bse.2006.01.011.
Naydenov K.D., Mladenov I., Alexandrov A., Naydenov M.K., Gyuleva V., Goudiaby V., Nikolić B., Kamary S. (2015). Patterns of genetic diversity resulting from bottlenecks in European black pine, with implications on local genetic conservation and management practices in Bulgaria. European Journal of Forest Research 134: 669–681. http://dx.doi.org/10.1007/s10342-015-0881-3.
Petit R.J., El Mousadik A., Pons O. (1998). Identifying populations for conservation on the basis of genetic markers. Conservation Biology 12: 844–855. http://dx.doi.org/10.1111/j.1523-1739.1998.96489.x.
Petković K. (1987). Geologija Srbije. [Geology of Serbia]. Zavod za regionalnu geologiju i paleontologiju rudarsko-geološkog fakulteta, Univerzitet u Beogradu. [In Serbian].
Slatkin M. (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462.
Šarac Z., Bojović S., Nikolić B., Tešević V., Đorđević I., Marin P.D. (2013). Chemotaxonomic significance of the terpene composition in natural populations of Pinus nigra J.F. Arnold from Serbia. Chemistry and Biodiversity 10: 1507–1520. http://dx.doi.org/10.1002/cbdv.201200416.
Šarac Z., Aleksić J.M., Dodoš T., Rajčević N., Bojović S., Marin P.D. (2015). Cross-species amplification of nuclear EST-microsatellites developed for other Pinus species in Pinus nigra. Genetika 47: 205–217. http://dx.doi.org/10.2298/GENSR1501205S.
Soto A., Robledo-Arnuncio J.J., González-Martínez S.C., Smouse P.E., Alía R. (2010). Climatic niche and neutral genetic diversity of the six Iberian pine species: a retrospective and prospective view. Molecular Ecology 19: 1396–1409. http://dx.doi.org/10.1111/j.1365-294X.2010.04571.x.
Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. http://dx.doi.org/10.1093/molbev/msr121.
Tian S., López-Pujol J., Wang H.-W., Ge S., Zhang Z.-Y. (2010). Molecular evidence for glacial expansion and interglacial retreat during Quaternary climatic changes in a montane temperate pine (Pinus kwangtungensis Chun ex Tsiang) in southern China. Plant Systematics and Evolution 284: 219–229. http://dx.doi.org/10.1007/s00606-009-0246-9.
Vendramin G.G., Lelli L., Rossi P., Morgante M. (1996). A set of primers for the 414 amplification of 20 chloroplast microsatellites in Pinaceae. Molecular Ecology 5: 595–598. http://dx.doi.org/10.1111/j.1365-294X.1996.tb00353.x.
Vidaković M. (1991). Conifers. Morphology and variation. Grafički Zavod Hrvatske, Zagreb.
Weir B.S. (1996). Genetic data analysis II: Methods for discrete population genetic data. Sinauer Associates, Sunderland, MA, USA.
Total of 39 references.