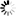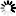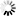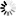Genetic diversity and differentiation of the riparian relict tree Pterocarya fraxinifolia (Juglandaceae) along altitudinal gradients in the Hyrcanian forest (Iran)
Yousefzadeh H., Rajaei R., Jasińska A., Walas Ł., Fragnière Y., Kozlowski G. (2018). Genetic diversity and differentiation of the riparian relict tree Pterocarya fraxinifolia (Juglandaceae) along altitudinal gradients in the Hyrcanian forest (Iran). Silva Fennica vol. 52 no. 5 article id 10000. https://doi.org/10.14214/sf.10000
Highlights
- The Caucasian wingnut (Pterocarya fraxinifolia Spach) is an emblematic and relict riparian tree with limited distribution in Hyrcanian forest which investigating its genetic population structure and diversity along altitudinal gradients, and migration patterns are novel
- We concluded that rivers are the main seed dispersal vector among P. fraxinifolia populations and there was no trend from upstream to downstream
- The high level of gene flow and uniform genetic diversity along each river system suggest the “classical” metapopulation structure of the species.
Abstract
Riparian trees, especially relict trees, are attractive and important for research to understand both past and recent biogeographical and evolutionary processes. Our work is the first study to elucidate the genetic diversity and spatial genetic structure of the canopy-dominating riparian Pterocarya fraxinifolia (Juglandaceae) along two altitudinal gradients in different river systems of the Hyrcanian forest, which is one of the most important refugium of relict trees in Western Eurasia. Altitudinal gradients were chosen along two river systems at 100, 400 and 900 m a.s.l. Leaf samples were collected from 116 trees, and the genetic diversity was evaluated with eight SSR markers. Overall, 39 alleles were identified for all of the populations studied. The observed heterozygosity (Ho) varied from 0.79 to 0.87 (with a mean of 0.83). The results of the AMOVA analysis indicated that the variation within populations was 88%, whereas the variation among populations was 12% for all of the gradients. A structure analysis indicated that 93% of the trees were grouped in the same gradient. The genetic distance based on Fst confirmed the structure result and indicated a high rate of gene flow among the investigated populations. Based on high gene flow (low differentiation of the population along the same river) and the clearly distinct genetic structure of the investigated gradients, it can be concluded that rivers are the main seed dispersal vector among P. fraxinifolia populations. The genetic diversity of P. fraxinifolia did not show any trend from upstream to downstream. The high level of gene flow and uniform genetic diversity along each river suggest the “classical” metapopulation structure of the species.
Keywords
plant variation;
Caucasian wingnut;
population genetics;
refugium;
riparian forest;
river system
-
Yousefzadeh,
Department of Environmental Science, Faculty of Natural Resources and Marine Sciences, Tarbiat Modares University, Noor, Mazandaran, Iran
E-mail
h.yousefzadeh@modares.ac.ir

- Rajaei, Department of Forestry, Faculty of Natural Resources and Marine Sciences, Tarbiat Modares University, Noor, Mazandaran, Iran E-mail r.rajaei@modares.ac.ir
- Jasińska, Laboratory of Systematics and Geography, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, PL-62-035 Kornik, Poland E-mail jasiak9@wp.pl
- Walas, Laboratory of Systematics and Geography, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, PL-62-035 Kornik, Poland E-mail lukaswalas@wp.pl
- Fragnière, Department of Biology and Botanic Garden, University of Fribourg, Chemin du Musée 10, CH-1700 Fribourg, Switzerland E-mail yann.fragniere@unifr.ch
- Kozlowski, Department of Biology and Botanic Garden, University of Fribourg, Chemin du Musée 10, CH-1700 Fribourg, Switzerland; Natural History Museum Fribourg, Chemin du Musée 6, CH-1700 Fribourg, Switzerland E-mail gregor.kozlowski@unifr.ch
Received 4 May 2018 Accepted 23 November 2018 Published 14 December 2018
Views 151765
Available at https://doi.org/10.14214/sf.10000 | Download PDF

Supplementary Files
1 Introduction
Riparian forests, which are characterized by challenging environmental conditions, are very important due to their high level of biodiversity, which supplies a multitude of environmental, economic and aesthetic benefits (Sakio and Tamura 2008). The major threats for these sensible habitats lies in human disturbance, particularly due to clearing for agriculture and urban development, grazing, invasive species and disrupted river ecosystem through artificial barriers such as dam constructions (Imbert and Lefevre 2003; Smulders et al. 2008). As a consequence, it has been estimated that up to 99% of the riparian forests in Europe have disappeared (Lefevre et al. 1998; Hughes and Rood 2003). Currently, there is increasing interest in restoring riparian habitats (Hughes et al. 2005). Understanding the genetic structure of the population and genetic differentiation helps in the management and restoration of endangered species (Holderegger and Wagner 2000; Yang et al. 2015) and, more specifically, of riparian forest trees (Wei et al. 2015).
During the last half century, an additional threat has occurred from accelerated climate change (Leadley et al. 2010). Relict plants, especially relict trees, which have been able to survive impressive environmental changes over millions of years, are of great concern for their valuable contribution to improving the understanding of both past and recent biogeographical and evolutionary processes (Kozlowski and Gratzfeld 2013). An effective way to deal with the damage caused by climate change is to preserve the genetic diversity within a population of a species (Ehlers et al. 2008; Pauls et al. 2013). Species can react to global climate change by adapting and persisting in new conditions (Hoffmann and Sgro 2011) or by shifting their habitat or range (Chen et al. 2011). Without adaptation or distribution shift capacities, these fast climatic changes can lead to extinction (Dawson et al. 2011). Thus, preserving the genetic diversity within a species is very important for its long-term survival (Pauls et al. 2013). Climate relicts are the remnants of past populations that have become isolated and/or fragmented by climate-driven range shifts that preserved ecological and evolutionary histories (Ian Milne 2006; Woolbright 2014). Relict trees and their forests have the potential to enhance our understanding of how long-term environmental change affects species’ interactions, Forest community composition and disassembly as well as the services that ecosystems provide. They enable the study of changes and interactions that occurred inside communities during a time span of millions of years through climate change and habitat fragmentation (Lyons 2016). Many relict woody plants are confined to multiple refugia (Kozlowski and Gratzfeld 2013). This can be caused by limited seed dispersal abilities or the presence of unsuitable habitats surrounding refugia that could prevent the migration of taxa and range expansion. Due to geographical isolation, the gene flow between refugial areas would have been negligible during interglacials and glacial periods, and mutations would have expanded little from their place of origin (Hewitt 1996, 2001). Moreover, isolated relict populations would have been exposed to bottleneck situations due to their small size (Hampe and Arroyo 2002). However, comparatively little is known about the phylogeography and genetic structure of the plants confined to multiple refugial regions.
For our study, we chose the Caucasian wingnut (Pterocarya fraxinifolia Spach, Juglandaceae), which is a riparian tree. This is the only species of the genus occurring in Western Eurasia. Most extant species of the wingnut genus (P. hupehensis Skan, P. macroptera Batalin, P. stenoptera C. DC, P. tonkinensis (Franch.) Dode and P. rhoifolia Siebold & Zucc.) are distributed in eastern Asia (Zheng-Yi and Raven 2003). Pterocarya is an ancient relict genus, with the oldest fossil fruits dating to the Eocene-Oligocene in North America and Eurasia (Manchester and Dilcher 1982). In Europe, Pterocarya fossils dating to the Pleistocene interglacial periods have been found in Germany, Poland, Lithuania, Ukraine, France, and Italy (Binka et al. 2003; Ravazzi et al. 2005; Guiter et al. 2008). However, wingnuts went extinct in western Eurasia, except in the Euxino-Hyrcanian Province (Takhtadjan 1986; Zohary 1973) and its small southern outposts (Boratyńska and Boratyński 1975). The Hyrcanian forests, where P. fraxinifolia occurs, are recognized as one of the most biodiverse ecosystems and a reservoir of relict woody species with high strategic value for the conservation and maintenance of ecosystem services (Browicz 1988, 1997; Maharramova 2016; Maharramova et al. 2018).
Pterocarya fraxinifolia is a thermophilus deciduous tree species with a lifespan of 200–250 years that grows along watercourses in lowland and ascending along riparian forests in the mountains up to 1700 m a.s.l. It is a synchronously monoecious species with many-flowered pistillate and staminate catkins (Iljinskaya 1970) that bloom between mid-April and mid-May, produces wind-dispersed pollen in abundance and forms winged nutlike drupes that are dispersed by both wind and water. Vegetative reproduction in this species is common and occurs through root sprouting (Gulisashvili 1961). In the South Caucasus, P. fraxinifolia occurs mainly in the Colchis on the eastern coast of the Black Sea, in the Hyrcanian region on the southern coast of the Caspian Sea, and on the southern slopes of the Greater Caucasus mountain range and the adjacent valleys (Browicz 1989, 1997). Additional populations can be found in Anatolia (Avsar and Ok 2004) and the central Zagros Mountains (Akhani and Salimian 2003). Pterocarya fraxinifolia commonly occurs in lowland areas, which has been frequently converted for agricultural, industrial and regional maintenance purposes (Biltekin et al. 2014; Scharnweber et al. 2007). Due to this decline, P. fraxinifolia, along with other relict and endemic taxa, is currently under protection in national parks and state reserves in Azerbaijan, Georgia and Iran. However, future changes toward drier climates might further endanger the species (Denk et al. 2001). Agricultural alteration of the natural landscape and low genetic diversity may particularly restrict the adaptation of the species to a changing environment (McLaughlin et al. 2002).
Information on the patterns of genetic diversity in relict trees can help elucidate how these species adapt to climate changes in the long term (Holderegger and Wagner 2000; Yang et al. 2015). To date, only a few genetic studies exist on the genus Petrocarya (Sugahara et al. 2017), and only two studies have investigated the phylogeny and phylogeography of P. fraxinifolia in the Transcaucasia and Hyrcanian regions (Maharramova 2016; Mostajeran et al. 2017).
Referring to Ritland’s unidirectional diversity hypothesis (Ritland 1989) (which concerns that genetic flow is relative to the unidirectional flow of abiotic environmental influences, particularly stream flow) we employed nuclear microsatellite markers to investigate the genetic diversity and genetic structure of P. fraxinifolia in a natural river system of Northern Iran (Alborz Mountains). In recent years, numerous studies on genetic diversity along an altitudinal gradient have been conducted using molecular markers (McMahon et al. 2011). However, in our study, we intend to (i) evaluate the differences in the genetic diversity of P. fraxinifolia populations along altitudinal gradients of different river systems, (ii) evaluate the differences between the rate of gene flow among the populations along the same river compared with different river systems, and (iii) discuss the implications of the results for the conservation of P. fraxinifolia and the restoration of its riparian forests.
2 Materials and methods
2.2 Sampling of plant material, DNA extraction and SSR amplification
Two altitudinal gradients were chosen along valleys of Talar and Tajan rivers (Fig. 1), in the center of the Hyrcanian forest, north of Iran (Table 1). Three populations have been selected along each river, using altitudinal gradient at approximately 100, 400 and 900 m a.s.l. In every population, 18–20 trees, separated by distance of at least 20 m from each other, were collected (leaf samples). Before DNA extraction, the leaves were frozen in liquid nitrogen and ground to a fine powder using a mortar and pestle. The total genomic DNA was isolated from the ground powder using a protocol modified from Janfaza et al. (2017), and based on the method of Murray and Thompson (1980). Eight nuclear SSR (Simple Sequence Repeats markers) were selected, and PCR and cycle amplifications were performed according to Maharramova (2016). The PCR products were separated by 8% polyacrylamide gel electrophoresis (PAGE) and detected by silver staining. Fragment analysis was conducted using the Gel-Pro Analyzer software.
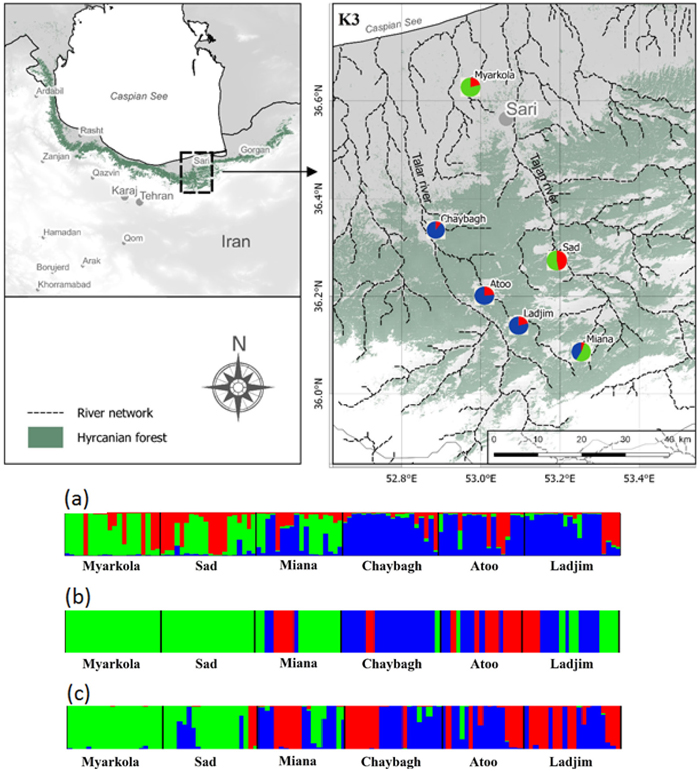
Fig. 1. Bayesian clustering results for Pterocarya fraxinifolia in the Hyrcanian region (N = 116) estimated along two altitudinal gradients from K = 3 and an associated map of the mean membership probabilities per site from STRUCTURE. Each individual is represented by a vertical bar partitioned into K segments, which represents the amount of ancestry of its genome corresponding to K clusters. Visualization was improved by sorting the genotypes by site. Gradient 1: Myarkola, Sad, and Miana; gradient 2: Chaybagh, Atoo, and Ladjim. (a) Output from STRUCTURE. (b) Output from BAPS. (c) Results of DAPC analysis in R.
| Table 1. Geographical characteristics and sample size of investigated populations in two gradients along two river systems. | |||||
| Population name | Latitude (UTM) | Longitude (UTM) | Altitude (m a.s.l.) | Sample size | |
| Tajan river (gradient 1) | Myarkola | 672536.3 | 4051845.08 | 50–60 | 20 |
| Sad | 697694.9 | 4017597.8 | 350–400 | 20 | |
| Miana | 703567.39 | 3996839.29 | 900–970 | 18 | |
| Talar river (gradient 2) | Chaybagh | 669888.3 | 4023492.3 | 200–250 | 20 |
| Atoo | 681346.47 | 4008915.14 | 400–450 | 18 | |
| Ladjim | 686691.74 | 4013147.26 | 900–950 | 20 | |
2.2 Statistical analysis
The presence of null alleles was estimated by Microchecker v2.2.3 (Van Oosterhout 2004). The number of observed (Na) and effective alleles (Ne), observed and expected heterozygosity, inbreeding index, clonal structure and gene flow among the populations were calculated by the GenAlEx software (Peakall and Smouse 2006). Clonal structure in studied populations was calculated using genotype diversity according to the formula R = (G − 1)/(N − 1), where G is the number of total genotypes detected and N is the number of individuals collected along both river gradients (Dering et al. 2017).
Genetic differentiation among the populations was estimated by an analysis of molecular variance (AMOVA). The average differentiation coefficients (Fst), gene flow (Nm) and pair wise population assignment test were calculated using GenAlEx v6.5. Allelic richness (Ar) was calculated using the rarefaction method with the HP-Rare software (Kalinowski 2004, 2005). The population structure and assignment of trees to the populations were performed using STRUCTURE v2.3.3. (Pritchard et al. 2000). The allele frequencies were correlated between populations with the admixture model, and the algorithm was run with 50 000 burn-in Markov Chain Monte Carlo (MCMC) iterations and 50 0000 MCMC post burn-in repetitions for the parameter estimation. The structure was run in three replicates with assumed K from 2 to 8 (Supplementary files S1 and S2). The structure harvester’ online program was used to determine the appropriate K value based on both the LnP (D) and Evanno’s ΔK (Evanno et al. 2005). Additionally, it was used BAPS (Bayesian Analysis of Population Structure) where analyze was conducted with 1000 iterations (Corander et al. 2006, 2008; Cheng et al. 2011). DAPC (Discriminant Analysis of Prinicipal Components) from adegenet package in R (Jombart 2008; Jombard and Ahmed 2011) was used complementary to Bayesian clustering methods from STRUCTURE and BAPS. Number of K was 3 and number of used PCA was 7.
3 Results
3.1 Microsatellite variation and genetic diversity
Null alleles were detected in five populations at locus 2 and in only one population at locus 5. Thus, the six remaining SSR loci were determined to be a proper set to assess the genetic diversity. Overall, 39 alleles were identified for all of the studied populations in two gradients. For both the number of observed alleles (Na) and effective alleles (Ne), the maximum and minimum values were related to Chaybagh and Myarkola populations (Table 2). The observed heterozygosity (Ho) varied from 0.79 (Chaybagh) to 0.87 (Atoo), with a mean of 0.83 for all populations. Except for the Myarkola population, the Ho was higher than He.
| Table 2. Genetic variability within two gradients of Pterocarya fraxinifolia populations. | |||||||
| Elevation | Population | Effective number of alleles (Ne) | Observed heterozygosity (Ho) | Expected heterozygosity (He) | Allelic richness (Ar) | Private allelic richness (PAr) | |
| Gradient 1 | Low | Myarkola | 5.7 ± 0.73 | 0.78 ± 0.1 | 0.807 ± 0.028 | 4.06 | 0.99 |
| Medium | Sad | 4.7 ± 0.36 | 0.85 ± 0.067 | 0.78 ± 0.02 | 3.8 | 0.4 | |
| High | Miana | 5.1 ± 0.44 | 0.86 ± 0.07 | 0.79 ± 0.02 | 3.9 | 0.38 | |
| Gradient 2 | Low | Chaybagh | 3.72 ± 0.23 | 0.79 ± 0.11 | 0.726 ± 0.018 | 3.3 | 0.1 |
| Medium | Atoo | 4.27 ± 0.51 | 0.87 ± 0.09 | 0.751 ± 0.025 | 3.5 | 0.2 | |
| High | Ladjim | 4.25 ± 0.69 | 0.86 ± 0.11 | 0.730 ± 0.043 | 3.4 | 0.14 | |
3.2 Genetic differentiation, population structure and clonality
The results of the AMOVA analysis indicated that variation within the populations was 91%, 93% and 88%, whereas variation among the populations was 9% and 7% and 12%. Gene flow were 5.13, 6.6 and 3.77 (as the average numbers of migrants per generation) for gradient 1, gradient 2 and all populations (the two gradients mixed), respectively (Table 3).
| Table 3. Analyses of molecular variance (AMOVA) for two gradients of Pterocarya fraxinifolia from Hyrcanian forest using SSR markers. | ||||||||||
| AMOVA component | AMOVA genetic distance option | |||||||||
| S.O.V | d.f | SS | MS | Est. Var. | % of Variance | PhiPT | Rst | Fst | Nm | |
| Gradient 1 | Among Pops | 2 | 27.76 | 13.8 | 0.45 | 9% | 0.086** | 0.049** | 0.046** | 5.13 |
| Within Pops | 55 | 284.95 | 5.18 | 5.18 | 91% | |||||
| Total | 57 | 312.72 | 5.63 | 100% | ||||||
| Gradient 2 | Among Pops | 2 | 22.13 | 11.06 | 0.34 | 7 | 0.075** | 0.052** | 0.036** | 6.6 |
| Within Pops | 55 | 237.68 | 4.32 | 4.32 | 93 | |||||
| Total | 57 | 259.82 | 4.67 | 100 | ||||||
| Total | Among Pops | 5 | 85.1 | 17 | 0.63 | 12 | 0.11** | 0.037** | 0.062** | 3.77 |
| Within | 110 | 521.3 | 4.7 | 88 | ||||||
| Total | 115 | 606.4 | 4.7 | 5.37 | 100 | |||||
| Statistics include sums of squared deviations (SS); mean squared deviations (MS), variance component estimates (Est. Var.). ** – p-value < 0.01. | ||||||||||
The structure analysis was conducted three times based on each altitudinal gradient (river system) and then for all populations together (Fig. 1 and Suppl. file S1). The delta K was greatest for K = 3, which suggests the presence of three main groups in the two gradients (Suppl. file S2). The structure analysis indicated that more than 93% of the trees were grouped based on the gradient but did not group the trees based on the altitude in each gradient. However, intermixing of the colors across the altitude were also observed. In the first gradient, the trees represented 24.66%, 61.61% and 13.73% in groups 1, 2 and 3, respectively, whereas 81.81% of the trees in gradient 2 were located solely in group 3. The heterogeneity in gradient 1 was higher than that in the populations of gradient 2. Only 6.03% (7 trees) were placed in the admixture due to a membership coefficient of less than 0.55. Results from BAPS and DAPC are similar to result obtained in STRUCTURE (Fig. 1), but indicate a higher heterogeneity in gradient 2. It can be seen spatial pattern of genetic structure: majority of individuals from Myarkola and Sad form one genetic cluster, while populations from gradient 2 are dominated by another two clusters. Results of DAPC with populations used as a clusters show a clear difference between the two gradinets (Fig. 2), however, Miana shows similarity to both gradients.
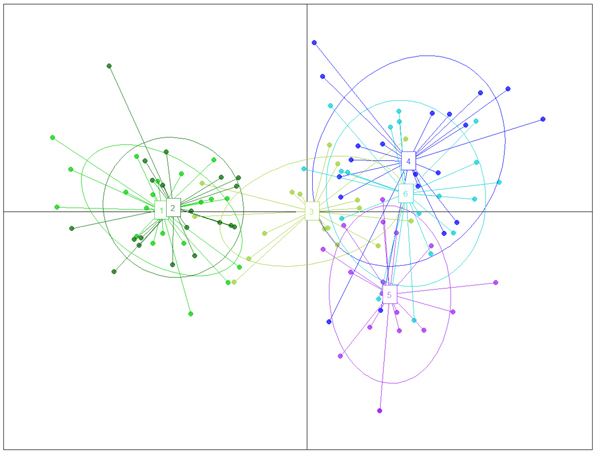
Fig. 2. The result of DAPC analysis with populations used as clusters: 1 – Myarkola, 2 – Sad, 3 – Miana, 4 – Chaybagh, 5 – Atoo, 6 – Ladjim. Green colours indicated populations from gradient 1, blue and purple – populations from gradient 2.
The assignment proportions of each individual to the population are surprising since a total of 94% of individuals are clustered in the populations originated from themselves (Fig. 3 and Table 4). Of the six populations, only the individuals from the Sad and Miana populations had 100% assignment success.
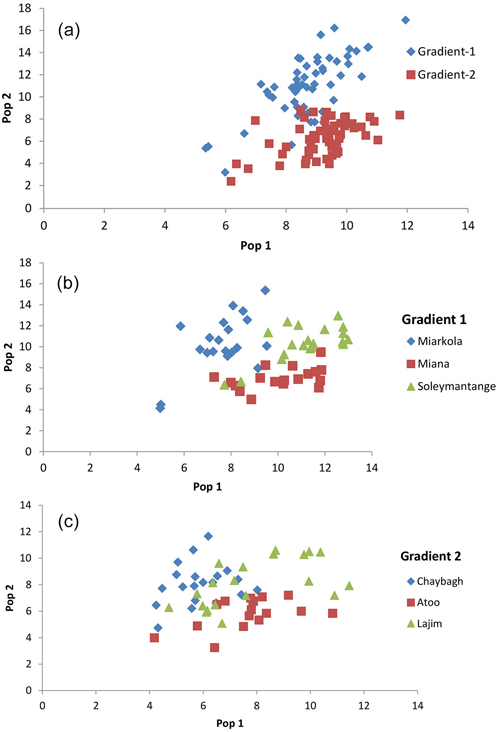
Fig. 3. Result of the population assignment. (a) Pairwise population assignment graphs of gradient 1 vs. gradient 2. (b) Assignment of the populations of gradient 1. (c) Assignment of the populations of gradient 2.
| Table 4. Summary of population assignment outcomes to “self” or “other” population. | ||||||
| Pop | Gradient 1 Pops | Gradient 2 Pops | Total | Percent | ||
| All populations* | Self Pop | 52 | 57 | 109 | 94% | |
| Other Pop | 6 | 1 | 7 | 6% | ||
| Pop | Myarkola | Sad | Miana | Total | Percent | |
| Gradient 1 | Self Pop | 17 | 20 | 18 | 55 | 95% |
| Other Pop | 3 | 0 | 0 | 3 | 5% | |
| Pop | Chaybagh | Atoo | Ladjim | Total | Percent | |
| Gradient 2 | Self Pop | 18 | 16 | 16 | 50 | 86% |
| Other Pop | 2 | 2 | 4 | 8 | 14% | |
| * – All population in each gradient grouped as one population. | ||||||
The genetic distance based on Fst also confirmed the structure result, where the Miana population from gradient 1 had the lowest genetic distance with the Ladjim population from gradient 2. Additionally, a high rate of gene flow was observed among the investigated populations (Table 5).
| Table 5. Pairwise population gene flow (Nm) values (below diagonal) and pairwise population genetic distance based on Fst values (above diagonal). | ||||
| Populations | Myarkola | Sad | Miana | |
| Gradient 1 | Myarkola | 0.000 | 0.058 | 0.025 |
| Sad | 9.88 | 0.000 | 4.343 | |
| Miana | 4.343 | 0.054 | 0.000 | |
| Populations | Chaybagh | Atoo | Ladjim | |
| Gradient 2 | Chaybagh | 0.000 | 0.041 | 0.026 |
| Atoo | 5.856 | 0.000 | 0.043 | |
| Ladjim | 9.310 | 5.564 | 0.000 | |
| Nm – the product of the effective population number and rate of migration among populations. | ||||
Number of genotypes was the equal as number of individuals in all investigated populations except Atoo, where number of genotypes was one smaller than the number of individuals. This means that the applied methodology of the collection allowed to capture the entire (or the majority) of the genetic variability of selected populations regardless of asexual reproduction which is known to occur in P. fraxinifolia through root sprouting (Gulisashvili 1961).
3.3 Migration pattern
The results of this study show that the genetic migration and heterozygosity of P. fraxinifolia did not follow a regular pattern. The highest gene flow was observed between the upper (Miana and Ladjim) and lower (Myarkola and Atoo) populations. Heterozygosity (He) was approximately equal in all three populations (high, middle and lowest elevation). The gene flow between the populations within each river was higher than that between populations belonging to the from different rivers (Fig. 4), except for the Miana population from the Tajan river, which seemed to have a high genetic flow due to the small geographic distance from the Ladjim population of the second river (Fig. 4).
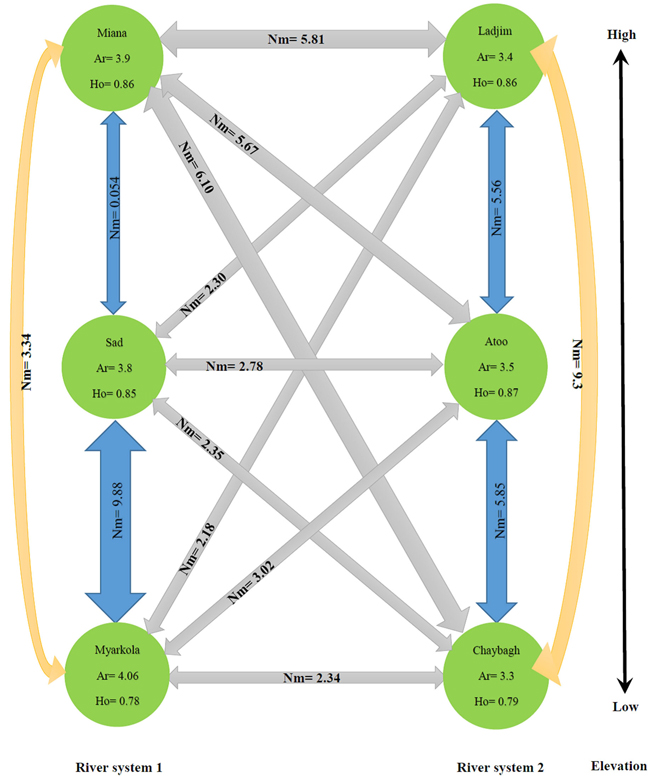
Fig. 4. Migration pattern of Pterocarya fraxinifolia within and among the river populations.
4 Discussion
Our work is the first study to elucidate the genetic diversity and spatial genetic structure of P. fraxinifolia along two altitudinal gradients in different river systems of the Hyrcanian forest, which is one of the most important refugium of the relict trees in Western Eurasia.
A high level of genetic diversity was detected for P. fraxinifolia in both studied gradients. The genetic diversity was higher within populations than it was among populations. Thus, P. fraxinifolia is an exception among riparian plant species because in the majority of these plants, the genetic diversity was reported to be low to medium (DeWoody et al. 2004; Prentis and Mather 2008; Russell et al. 1999). Interestingly, Maharramova (2016) reported that natural populations of the species show low-to-intermediate levels of genetic diversity and a significant decrease of diversity from Hyrcan to Colchis. However, in her study covering the whole distribution area of the species, the distance between populations was much larger.
The results revealed that, the populations belonging to the same gradient showed higher similarity, especially along gradient 2. The situation was slightly different in gradient 1 because of the construction of a dam between the highest population (Miana) and populations at lower altitudes (Sad and Myarkola). This artificial barrier probably disconnected the gene flow (occurring via seeds) from higher to lower populations. However, our study delivers contrary conclusions to those of Werth and Scheidegger (2014) who show that the construction of dams and dikes has no effect on heterozygosity of downstream populations. The age of the sampled trees is higher than the age of the dam (27 years). Thus, if we want to examine the negative effect of dam construction, we should do the same study at the seedling level.
Our study supports the results of Maharramova (2016), who reports the absence of significant inbreeding and genetic drift, and confirms that the high genetic diversity among populations of P. fraxinifolia results from outcrossing and wind-pollination of this tree species, which allows long-distance gene flow (Heuertz et. 2003).
The significant structure along two gradients indicates that the gene flow among P. fraxinifolia populations is higher along the same river than between populations belonging to different river systems. Isolation by distance and differentiation along a river system, which was reported previously for P. fraxinifolia by Maharramova (2016) in the Hyrcanian forest, is related to higher forest density, special topography (high mountain between the neighboring river valleys) or other barriers that limit the wind dispersal of pollen/seeds. Rivers play a fundamental role in maintaining gene flow between populations of riparian trees. Pterocarya fraxinifolia and other riparian trees grow right at the waterfront and drop seeds directly into the water. However, we detected gene flow between populations belonging to two different gradients. The populations of Miana and Ladjim (the populations with the highest altitude in two gradients) are growing relatively close together in terms of geographical distance and have a greater chance for gene-flow by pollen than other populations. The structure analysis confirmed the possibility of gene flow and mixing between these two populations (Fig. 1). BAPS and DAPC analysis also indicated that Miana show a relatively high similarity to the population from gradient 2 (Figs. 2 and 3).
Genetic differentiation of populations along both gradients, and based on Fst, was low, similar to that of other riparian species, such as Hymenocallis coronaria (J. Le Conte) Kunth (Markwith and Scanlon 2006; Kudoh and Whigham 1997), Hibiscus moscheutos L. (Van Der Meer and Jacquemyn 2015), Populus nigra L. (Imbert and Lefevre 2003), Boltonia decurrens (Torr. & A. Gray) Alph. Wood (DeWoody et al 2004) and Saxifraga granulata L. (Van Der Meer and Jacquemyn 2015). These values were reported to be relatively high for some riparian species (Lundqvist and Andersson 2001; Liu et al. 2006). Many factors, such as life form, geographical range and breeding system, have an impact on the proportion of population diversity and structure (Elistrand 2016; Chombe et al. 2017; Mosner et al. 2017).
The high genetic diversity versus low Fst of P. fraxinifolia was previously reported for other relict species as Michelia coriacea H.T. Chang & B.L. Chen (Zhao et al. 2012) and Zelkova carpinifolia (Pall.) K. Koch (Maharramova et al. 2015). One reason of this can be a high gene flow (Total Nm = 3.77) among the investigated populations. Van Rossum and Prentice (2004) demonstrated that, such pattern may also result of past events. Fossil evidence indicates that P. faxinifolia was widespread throughout the Hyrcanian forest and in many other regions of Iran and Transcaucasia during Pleistocene (Leroy et al. 2013) and populations of that species have been presumably naturally fragmented for approximately 10 000–7000 years (Akhani et al. 2010). Based on the low differentiation of the population along the river (high gene flow) and the clearly distinct genetic structure of the investigated gradients, it can be concluded that rivers are the main seed dispersal. As in other riparian plant species (Soons et al. 2008; James et al. 2013), other ways for gene-flow (e.g., pollen or vegetative propagules) play a less important role.
The high genetic diversity and high rate of gene flow among populations along a rivers might be the main reasons for the long-term survival of P. fraxinifolia in the Hyrcanian Forest. It was previously demonstrated that species with higher gene flow and genetic diversity have higher phenotypic plasticity (Reed and Frankham 2003; Ghalambor et al 2007; Gruber et al. 2013). Moreover, multidirectional genetic diversity and allelic richness in all of the studied populations attests to sufficient gene flow and thus proves the genetic capacity of P. fraxinifolia to cope with environmental changes and global warming. In the case of the P. fraxinifolia population, growing along the Talar and Tajan rivers, we have not discovered a high level of clonality, typical for riparian species such as Alnus incana (L.) Moench (Dering et al. 2017), Populus nigra (Barsoum et al. 2004) or Populus euphratica Olivier, where in the most extreme situation a single genet can be determined (Sękiewicz, unpublished data). This occurs despite the severe degradation of the habitats of this species, especially in the plain region of the Hyrcanian region.
5 Conclusions
The migration pattern in P. fraxinifolia is explained by a classical metapopulation model along a river and a bidirectional gene flow among upper and lower populations. In the present study, we cannot find a changing trend in the genetic diversity of populations from upstream toward downstream.
References
Akhani H., Salimian M. (2003). An extant disjunct stand of Pterocarya fraxinifolia (Juglandaceae) in the central Zagros, W Iran. Willdenowia 33(1): 113–120. https://doi.org/10.3372/wi.33.33111.
Akhani H., Djamali M., Ghorbanalizadeh A., Ramezani E. (2010). Plant biodiversity of Hyrcanian relict forests, N Iran: an overview of the flora, vegetation, palaeoecology and conservation. Pakistan Journal of Botany 42: 231–258.
Avsar M.D., Ok T. (2004). A new distribution of Caucasian wingnut (Pterocarya fraxinifolia (Poiret) Spach) in the Kahramanmaras region, Turkey. Journal of Environmental Biology 25(1): 45–50.
Barsoum N., Muller E., Skot L. (2004). Variations in levels of clonality among Populus nigra L. stands of different ages. Evolutionary Ecology 18(5–6): 601–624. https://doi.org/10.1007/s10682-004-5146-4.
Biltekin D., Popescu S.M., Suc J.P., Quezel P., Jimenez-Moreno G., Yavuz N., Çağatay M.K. (2014). Anatolia: a long-time plant refuge area documented by pollen records over the last 23 million years. Review of Palaeobotany and Palynology 215: 1–22. https://doi.org/10.1016/j.revpalbo.2014.12.004.
Binka K., Nitychoruk J., Dzierzek J. (2003). Parrotia persica C.A.M. (Persian witch hazel, Persian ironwood) in the Mazovian (Holsteinian) Interglacial of Poland. Grana 42(4): 227–233. https://doi.org/10.1080/00173130310016220.
Boratynska K., Boratynski A. (1975). Geographical distribution of Pterocarya fraxinifolia Spach. Arboretum Kornickie 20: 131–138.
Browicz K. (1989). Chorology of the Euxinian and Hyrcanian element in the woody flora of Asia. Plant Systematics and Evolution 162(1–4): 305–314. https://doi.org/10.1007/BF00936923.
Browicz K. (1997). Chorology of trees and shrubs in south-west Asia and adjacent regions/ Phytogeographical analysis. Bogucki Wydawnictwo Naukowe, Poznan.
Chen I., Hill J.K., Ohlemüller R., Roy DB, Thomas C.D. (2011). Rapid range shifts of species associated with high levels of climate warming. Science 333(6045): 1024–1026. https://doi.org/10.1126/science.1206432.
Cheng L., Connor T.R., Aanensen D.M., Spratt B.G., Corander J. (2011). Bayesian semi-supervised classification of bacterial samples using MLST databases. BMC Bioinformatics 12(302). https://doi.org/10.1186/1471-2105-12-302.
Chombe D., Bekele E., Bryngelsson T., Teshome A., Geleta M. (2017). Genetic structure and relationships within and between cultivated and wild korarima [Aframomum corrorima (Braun) P.C.M. Jansen] in Ethiopia as revealed by simple sequence repeat (SSR) markers. BMC Genetics 18(72). https://doi.org/10.1186/s12863-017-0540-4.
Corander J., Marttinen P., Mäntyniemi S. (2006). Bayesian identification of stock mixtures from molecular marker data. Fishery Bulletin 104: 550–558.
Corander J., Marttinen P., Sirén J., Tang J. (2008). Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9(539). https://doi.org/10.1186/1471-2105-9-539.
Dawson T.P., Jackson S.T., House J.I, Prentice I.C., Mace G.M. (2011). Beyond predictions: biodiversity conservation in a changing climate. Science 332: 53–58.
Denk T., Frotzler N., Davitashvili V. (2001). Vegetational patterns and distribution of relict taxa in humid temperate forests and wetlands of Georgia (Transcaucasia). Biological Journal of the Linnean Society 72(2): 287–332. https://doi.org/10.1111/j.1095-8312.2001.tb01318.x.
Dering M., Latałowa M., Boratyńska K., Kosiński P., Adam Boratyńsk A. (2017). Could clonality contribute to the northern survival of grey alder [Alnus incana (L.) Moench] during the Last Glacial Maximum? Acta Societatis Botanicorum Poloniae 86(1): 3523.
DeWoody J., Nason J.D., Smith M. (2004). Inferring demographic processes from the genetic structure of a metapopulation of Boltonia decurrens (Asteraceae). Conservation Genetics 5(5): 603–617. https://doi.org/10.1007/s10592-004-1985-3.
Ehlers A., Worm B., Reusch T.B.H. (2008). Importance of genetic diversity in eelgrass Zostera marina for its resilience to global warming. Marine Ecology Progress Series 355: 1–7. https://doi.org/10.3354/meps07369.
Elistrand N.C. (2016). Gene Flow by Pollen : Implications for plant conservation genetics. Oikos 63(1): 77–86. https://doi.org/10.2307/3545517.
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14(8): 2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x.
Ghalambor C., McKay J., Carroll S., Reznick D. (2007). Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Functional Ecology 21(3): 394–407. https://doi.org/10.1111/j.1365-2435.2007.01283.x.
Gruber K., Schoning C., Otte M., Kinuthia W., Hasselmann M. (2013). Distinct subspecies or phenotypic plasticity? Genetic and morphological differentiation of mountain honey bees in East Africa. Ecology and Evolution 3(10): 3204–3218. https://doi.org/10.1002/ece3.711.
Guiter F., Andrieu-Ponel V., de Beaulieu J.L., Nicoud G., Ponel P., Blavoux B., Gandouin E. (2008). Palynostratigraphy of some Pleistocene deposits in the Western Alps: A review. Quaternary International 190(1): 10–25. https://doi.org/10.1016/j.quaint.2008.05.005.
Gulisashvili V.Z. (1961). Dendroflora Kavkaza. [Dendroflora of the Caucasus]. Vol. 2. 334 p.
Hampe A., Arroyo J. (2002). Recruitment and regeneration in populations of an endangered South Iberian Tertiary relict tree. Biological conservation 107(3): 263–271. https://doi.org/10.1016/S0006-3207(02)00061-7.
Heuertz M., Vekemans X., Hausman J.F., Palada M., Hardy O.J. (2003). Estimating seed vs. pollen dispersal from spatial genetic structure in the common ash. Molecular Ecology 12(9): 2483–2495. https://doi.org/10.1046/j.1365-294X.2003.01923.x
Hewitt G.M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society 58(3): 247–276. https://doi.org/10.1111/j.1095-8312.1996.tb01434.x.
Hewitt G.M. (2001). Speciation, hybrid zones and phylogeography – or seeing genes in space and time. Molecular Ecology 10(3): 537–549. https://doi.org/10.1046/j.1365-294x.2001.01202.x.
Hoffmann A.A, Sgro C.M. (2011). Climate change and evolutionary adaptation. Nature 470: 479–485. https://doi.org/10.1038/nature09670.
Holderegger R., Wagner H. (2000). Landscape genetics. Bioscience 4: 508–523.
Hughes F.M.R., Rood S.B. (2003). Allocation of river flows for restoration of floodplain forest ecosystems: a review of approaches and their applicability in Europe. Environmental Management 32(1): 12–33. https://doi.org/10.1007/s00267-003-2834-8.
Hughes F.M.R., Colston A., Mountford J.O. (2005). Restoring riparian ecosystems: the challenge of accommodating variability and designing restoration trajectories. Ecology and Society 10(1): art.12. https://doi.org/10.5751/ES-01292-100112.
Iljinskaya I.A. (1970). A monograph of the genus Pterocarya Kunth. Israel Program for Scientific Translations, Jerusalem. 117 p.
Imbert E., Lefevre F. (2003). Dispersal and gene flow of Populus nigra (Salicaceae) along a dynamic river system. Journal of Ecology 91(3): 447–456. https://doi.org/10.1046/j.1365-2745.2003.00772.x.
James E.A., Jordan R., Griffin P.C. (2013). Spatial genetic analysis of two polyploid macrophytes reveals high connectivity in a modified wetland. Freshwater Biology 58(10): 2102–2113. https://doi.org/10.1111/fwb.12194.
Janfaza S., Yousefzadeh H., Hosseini Nasr S.M., Botta R., Asadi Abkenar A., Marinon D.T. (2017). Genetic diversity of Castanea sativa an endangered species in the Hyrcanian forest. Silva Fennica 51(1) article 1705. https://doi.org/10.14214/sf.1705.
Jombart T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11): 1403–1405. https://doi.org/10.1093/bioinformatics/btn129.
Jombart T., Ahmed I. (2011). adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27(21): 3070–3071. https://doi.org/10.1093/bioinformatics/btr521.
Kalinowski S.T. (2004). Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conservation Genetics 5(4): 539–543. https://doi.org/10.1023/B:COGE.0000041021.91777.1a.
Kalinowski S.T. (2005). HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes 5(1): 187–189. https://doi.org/10.1111/j.1471-8286.2004.00845.x.
Kozlowski G., Gratzfeld J. (2013). Zelkova – an ancient tree. Global status and conservation action. Natural History Museum Fribourg.
Kudoh H., Whigham D.F. (1997). Microgeographic genetic structure and gene flow in Hibiscus moscheutos (Malvaceae) populations. Journal of Biogeography 84(9): 1285–1293. https://doi.org/10.2307/2446054.
Leadley P., Pereira H., Alkemade R., Fernandez-Manjarres J., Proenca V., Scharlemann J., Walpole M.J. (2010). Biodiversity scenarios: projections of 21st century change in biodiversity and associated ecosystem services. Secretariat of the Convention on Biological Diversity, Montreal.
Lefevre F., Legionnet A., de Vries S., Turok J. (1998). Strategies for the conservation of a pioneer tree species, Populus nigra L., in Europe. Genetics Selection Evolution 30: S181–S196.
Leroy S.A.G., Kakroodi A.A., Kroonenberg S., Lahijani H.K., Alimohammadian H., Nigarov A. (2013). Holocene vegetation history and sea level changes in the SE corner of the Caspian Sea: relevance to SW Asia climate. Quaternary Science Reviews 70: 28–47. https://doi.org/10.1016/j.quascirev.2013.03.004.
Liu Y., Wang Y., Huang H. (2006). High interpopulation genetic differentiation and unidirectional linear migration patterns in Myricaria laxiflora (Tamaricaceae), an endemic riparian plant in the three gorges valley of the Yangtze River. Journal of Biogeography 93: 206–215.
Lundqvist E., Andersson E. (2001). Genetic diversity in populations of plants with different breeding and dispersal strategies in a free-flowing boreal river system. Hereditas 135(1): 75–83. https://doi.org/10.1111/j.1601-5223.2001.00075.x.
Lyons K.S., Amatangelo K.L., Behrensmeyer A.K., Bercovici A., Blois J.L., Davis M., DiMichele W.A., Du A., Eronen J.T., Tyler Faith J., Graves G.R., Jud N., Labandeira C., Looy C.V., McGill B., Miller J.H., Patterson D., Pineda-Munoz S., Potts R., Riddle B., Terry R., Tóth A., Ulrich W., Villaseñor A., Wing S., Anderson H., Anderson J., Waller D., Gotelli N.J. (2016). Holocene shifts in the assembly of plant and animal communities implicate human impacts. Nature 529: 80–83. https://doi.org/10.1038/nature16447.
Maharramova E. (2016). Genetic diversity and population structure of the relict forest trees Zelkova carpinifolia (Ulmaceae) and Pterocarya fraxinifolia (Juglandaceae) in the South Caucasus. Doctoral dissertation. PhD thesis, Freie Universität Berlin.
Maharramova E., Huseynova I., Kolbaia S., Gruenstaeudl M., Borsch T., Muller L.A.H. (2018). Phylogeography and population genetics of the riparian relict tree Pterocarya fraxinifolia (Juglandaceae) in the South Caucasus. Systematics and Biodiversity 16(1): 14–27. https://doi.org/10.1080/14772000.2017.1333540.
Maharramova E.H., Safarov H.M., Kozlowski G., Borsch T., Muller L.A. (2015). Analysis of nuclear microsatellites reveals limited differentiation between Colchic and Hyrcanian populations of the wind-pollinated relict tree Zelkova carpinifolia (Ulmaceae). Journal of Biogeography 102: 119–128.
Manchester S.R., Dilcher D.L. (1982). Pterocaryoid fruits (Juglandaceae) in the Paleogene of North America and their evolutionary and biogeographic significance. Journal of Biogeography 69: 275–286.
Markwith S.H, Scanlon M.J. (2006). Characterization of six polymorphic microsatellite loci isolated from. Molecular Ecology Notes 6(1): 72–74. https://doi.org/10.1111/j.1471-8286.2005.01141.x.
McLaughlin J.F., Hellmann J.J, Boggs C.L., Ehrlich P.R. (2002). Climate change hastens population extinctions. Proceedings of the National Academy of Sciences 99(9): 6070–6074. https://doi.org/10.1073/pnas.052131199.
McMahon S.M., Harrison S.P., Armbruster W.S., Bartlein P.J., Beale C.M., Edwards M.E., Kattge J., Midgley G., Morin X., Prentice I.C. (2011). Improving assessment and modelling of climate change impacts on global terrestrial biodiversity. Trends in Ecology and Evolution 26(5): 249–259. https://doi.org/10.1016/j.tree.2011.02.012.
Milne R.I. (2006). Northern hemisphere plant disjunctions: a window on tertiary land bridges and climate change? Annals of Botany 98(3): 465–472. https://doi.org/10.1093/aob/mcl148.
Mosner E., Eimert K., Hüwe U., Ziegenhagen B., Janßen A., Leyer I. (2017). Revisiting the provenance delineation of a widespread shrub, Frangula alnus – the role of spatial, temporal and environmental patterns. Tree Genetics and Genomes 13(63). https://doi.org/10.1007/s11295-017-1142-z.
Mostajeran F., Yousefzadeh H., Davitashvili N., Kozlowski G., Akbarinia M. (2017). Phylogenetic relationships of Pterocarya (Juglandaceae) with an emphasis on the taxonomic status of Iranian populations using ITS and trnH-psbA sequence data. Plant Biosystems 151(6): 1012–1021. https://doi.org/10.1080/11263504.2016.1219416.
Murray M.G., Thmposon W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8(19): 4321–4325. https://doi.org/10.1093/nar/8.19.4321.
Pauls S.U., Nowak C., Balint M., Pfenninger M. (2013). The impact of global climate change on genetic diversity within populations and species. Molecular Ecology 22(4): 925–946. https://doi.org/10.1111/mec.12152.
Peakall R., Smouse P.E. (2006). GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Molecular Ecology Notes 6(1): 288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x.
Prentis P.J., Mather P.B. (2008). Fine-scale patterns of genetic variation indicate non-equilibrium gene frequency divergence in the stream lily, Helmholtzia glaberrima. Freshwater Biology 53(5): 973–980. https://doi.org/10.1111/j.1365-2427.2008.01953.x.
Pritchard J.K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Ravazzi C., Pini R., Breda M., Martinetto E., Muttoni G., Chiesa S., Confortini F., Egli R. (2005). The lacustrine deposits of Fornaci di Ranica (late Early Pleistocene, Italian Pre-Alps): stratigraphy, palaeoenvironment and geological evolution. Quaternary International 131(1): 35–58. https://doi.org/10.1016/j.quaint.2004.07.021.
Reed D.H., Frankham. R. (2003). Correlation between fitness and genetic diversity. Conservation Biology 17(1): 230–237. https://doi.org/10.1046/j.1523-1739.2003.01236.x.
Ritland K. (1989). Genetic differentiation, diversity, and inbreeding in the mountain Monkeyflower (Mimulus caespitosus) of the Washington Cascades. Canadian Journal of Botany 67(7): 2017–2024. https://doi.org/10.1139/b89-255.
Russell J.R., Weber J.C., Booth A., Powell W., Sotelo-Montest C., Dawson I.K. (1999). Genetic variation of Calycophyllum spruceanum in the Peruvian Amazon Basin, revealed by amplified fragment length polymorphism (AFLP) analysis. Molecular Ecology 8(2): 199–204. https://doi.org/10.1046/j.1365-294X.1999.00551.x.
Sakio H., Tamura T. (2008). Ecology of Riparian forests in Japan: disturbance, life history, and regeneration. Springer. https://doi.org/10.1007/978-4-431-76737-4.
Scharnweber T., Rietschel M., Manthey M. (2007). Degradation stages of the Hyrcanian forests in southern Azerbaijan. Archiv für Naturschutz und Landschafts forschung 46:133–156.
Smulders M.J.M., Cottrell J.E., Lefevre F., van der Schoot J., Arens P., Vosman B., Tabbener H.E., Grassi F., Fossati T., Castiglione S., Krystufek V., Fluch S., Burg K., Vornam B., Pohl A., Gebhardt K., Alba N., Agúndez D., Maestro C., Notivol E., Volosyanchuk R., Pospíšková M., Bordács S., Bovenschen J., van Dam B.C., Koelewijn H.P., Halfmaerten D., Ivens B., van Slycken J., Vanden Broeck A., Storme V., Boerjan W. (2008). Structure of the genetic diversity in black poplar (Populus nigra L.) populations across European river systems: consequences for conservation and restoration. Forest Ecology and Management 255(5–6): 1388–1399. https://doi.org/10.1016/j.foreco.2007.10.063.
Soons M.B., Van Der Vlugt C., Van Lith B., Heil G.W., Klaassen M. (2008). Small seed size increases the potential for dispersal of wetland plants by ducks. Journal of Ecology 96(4): 619–627. https://doi.org/10.1111/j.1365-2745.2008.01372.x.
Sugahara K., Kaneko Y., Sakaguchi S., Ito S., Yamanaka K., Sakio H., et al. (2017). Quaternary range-shift history of Japanese wingnut (Pterocarya rhoifolia) in the Japanese Archipelago evidenced from chloroplast DNA and ecological niche modeling. Journal of Forest Research 22(5): 282–293. https://doi.org/10.1080/13416979.2017.1351837.
Takhtajan A. (1986). Floristic regions of the world. University of California Press, Berkeley, CA.
Van Der Meer S., Jacquemyn H. (2015). Genetic diversity and spatial genetic structure of the grassland perennial Saxifraga granulata along two river systems. Plos One 10(6): e0130463. https://doi.org/10.1371/journal.pone.0130463.
Van Oosterhout C., Hutchinson W.F., Wills D.P.M., Shipley P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4(3): 535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x.
Van Rossum F., Prentice H.C. (2004). Structure of allozyme variation in Nordic Silene nutans (Caryophyllaceae): population size, geographical position and immigration history. Botanical Journal of the Linnean Society 81(3): 357–371. https://doi.org/10.1111/j.1095-8312.2003.00301.x.
Wei X., Meng H., Bao D., Jiang M. (2015). Gene flow and genetic structure of a mountain riparian tree species, Euptelea pleiospermum (Eupteleaceae): how important is the stream dendritic network? Tree Genetics and Genomes 11(64). https://doi.org/10.1007/s11295-015-0886-6.
Werth S., Scheidegger C. (2014). Gene flow within and between catchments in the threatened riparian plant Myricaria germanica. PLoS One 9(7): e994400. https://doi.org/10.1371/journal.pone.0099400.
Woolbright S.A., Whitham T.G., Gehring C.A., Allan G.J., Bailey J.K. (2014). Climate relicts and their associated communities as natural ecology and evolution laboratories. Trends in Ecology and Evolution 29(7): 406–416. https://doi.org/10.1016/j.tree.2014.05.003.
Yang Y.Q., Huang B.H., Yu Z.X., Liao P.C. (2015). Inferences of demographic history and fine-scale landscape genetics in Cycas panzhihuaensis and implications for its conservation. Tree Genetics and Genomes 11(78). https://doi.org/10.1007/s11295-015-0894-6.
Zhao X., Ma Y, Sun W., Wen X., Milne R. (2012). High genetic diversity and low differentiation of Michelia coriacea (Magnoliaceae), a critically endangered endemic in southeast Yunnan, China. The International Journal of Molecular Sciences 13(4): 4396–4411. https://doi.org/10.3390/ijms13044396.
Zheng-Yi W., Raven P.H. (2003). Pterocarya (Juglandaceae). Flora of China. 4: 280–282.
Zohary M. (1973). Geobotanical foundations of the Middle East. Vols. 1–2. Fischer, Stuttgart.
Total of 84 references.